 |
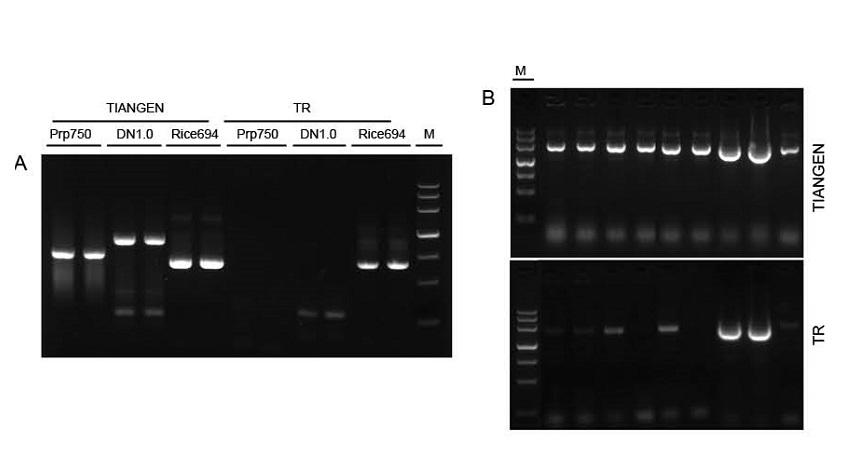
Figure 1. Templates from diff erent sources were amplified by TIANGEN Taq MasterMix II and the common Taq Mix from Supplier TR respectively to detect the stress resistance of the reagents. The results show that TIANGEN products can amplify the target fragments from crude genomic templates and bacterial culture, and the stress resistance is better than that of Supplier TR. A: Crude genomic template extracted by TIANGEN TIANcombi DNA Lyse&Det PCR Kit. Prp/DN: Crude extraction and detection of human blood samples. Rice: Crude extraction and detection of rice samples. B: Colony PCR. The PCR fragment is 700 bp. M: TIANGEN Marker III |
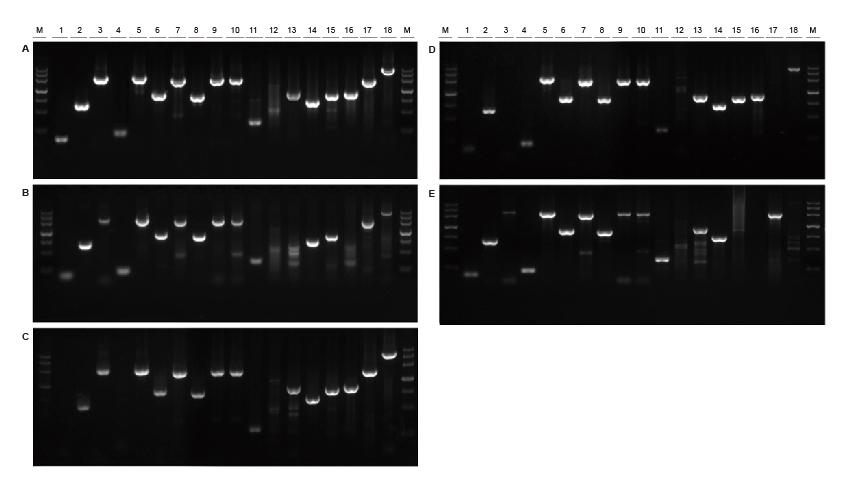 |
Good universality for templates from different sources and with different lengths Figure 2. Fragments of different sources and lengths were amplified using TIANGEN Taq MasterMix II (A) and ordinary Taq Mix of Supplier TK (B), Supplier TR (C), Supplier V (D) and Supplier G (E) respectively. The results show that the comprehensive performance of TIANGEN products is the best in terms of amplification capability, specificity and universality.M: TIANGEN Marker III1: Soybean genomic DNA template (120 bp); 2-3: Rice genomic DNA template (694 bp, 2258 bp); 4: Cotton genomic DNA template (200 bp); 5: Escherichia coli genomic DNA template (2298 bp); 6-7: Mouse genome DNA template (1 kb, 2 kb); 8-10: Rat genomic DNA template (1 kb, 2 kb, 2080 bp); 11-18: Human genome DNA template (300 bp, 448 bp (GC%: 74.8%), 1100 bp, 750 bp, 1000 bp, 1090 bp (GC%: 70.4%), 2 kb, 4 kb) |
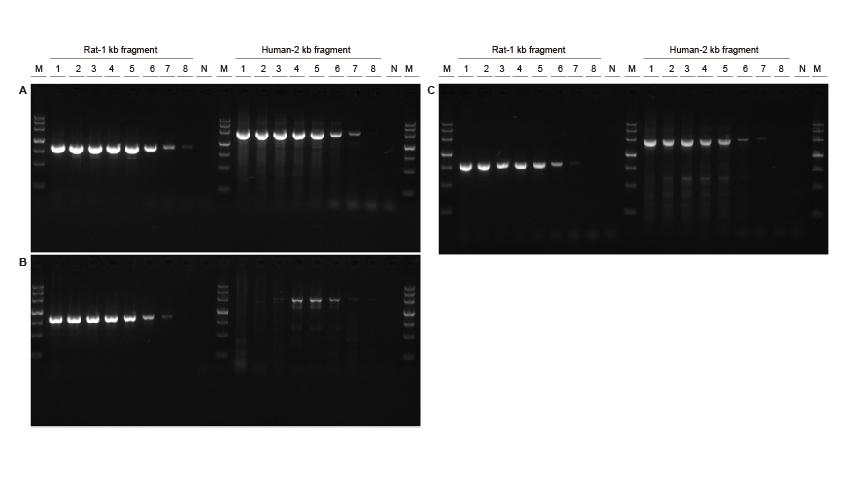 |
High sensitivity Figure 3. Different concentrations of rat and human DNA fragments were amplified using TIANGEN Taq MasterMix II (A), ordinary Taq Mix of Supplier V (B) and Supplier TK (C), respectively, to detect the amplification sensitivity. The results show that TIANGEN product could amplify the target fragment from the genome template as low as 0.01 ng, and its sensitivity is better than that of the products from Supplier V and TK.M: TIANGEN Marker III, N: NTCTemplate input 1-8: 200 ng, 100 ng, 50 ng, 20 ng, 10 ng, 1 ng, 0.1 ng, 0.01 ng. |
2×Taq PCR MasterMix Ⅱ
Features
■ High amplification efficiency: DNA fragments of different sizes (lower than 5 kb) and sources can be amplified efficiently.
■ High sensitivity: As low as 10 pg of target fragments can be amplified from genomic templates.
■ High stress resistance: For templates with high impurity content such as rough-extracted template/bacterial culture, the target fragment can be easily amplified. The polymerase activity will not be affected by repeated freezing and thawing.
■ Convenient for applications: The reaction system was prepared easily and quickly. The amplified fragment contains the 3′ end dA-overhang, which is convenient for TA cloning.
Specification
Type: Taq DNA polymerase
Sample: Purified/rough-extracted template/bacterial culture
Template: >10 pg
Fragment size: <5 kb
Applications: PCR amplification of DNA fragments, DNA labeling, primer extension, sequence determination, large-scale gene detection, semi-quantitative PCR experiments, detection of trace DNA, etc.
All the products can be customized for ODM/OEM. For details, please click Customized Service(ODM/OEM)
Experimental Example
FAQ
A-1 Template
■ The template contains protein impurities or Taq inhibitors, etc. ——Purify DNA template, remove protein impurities or extract template DNA with purification kits.
■ The denaturation of template is not complete ——Appropriately increase denaturation temperature and prolong denaturation time.
■ Template degradation ——Re-prepare the template.
A-2 Primer
■ Poor quality of primers ——Re-synthesize the primer.
■ Primer degradation ——Aliquot the high concentration primers into small volume for preservation. Avoid multiple freezing and thawing or long-term 4°C cryopreserved.
■ Inproper design of primers (e.g. primer length not sufficient, dimer formed between primers, etc.) -Redesign primers (avoid formation of primer dimer and secondary structure)
A-3 Mg2+concentration
■ Mg2+ concentration is too low ——Properly increase Mg2+ concentration: Optimize the Mg2+ concentration by a series of reactions from 1 mM to 3 mM with an interval of 0.5 mM to determine the optimal Mg2+ concentration for each template and primer.
A-4 Annealing temperature
■ The high annealing temperature affects the binding of primer and template. ——Reduce the annealing temperature and optimize the condition with a gradient of 2°C.
A-5 Extension time
■ Short extension time——Increase extension time.
Phenomena: Negative samples also show the target sequence bands.
A-1 Contamination of PCR
■ Cross contamination of target sequence or amplification products ——Carefully not to pipet the sample containing target sequence in the negative sample or spill them out of the centrifuge tube. The reagents or equipment should be autoclaved to eliminate existing nucleic acids, and the existence of contamination should be determined through negative control experiments.
■ Reagent contamination ——Aliquot the reagents and store at low temperature.
A-2 Primer
■ Mg2+ concentration is too low ——Properly increase Mg2+ concentration: Optimize the Mg2+ concentration by a series of reactions from 1 mM to 3 mM with an interval of 0.5 mM to determine the optimal Mg2+ concentration for each template and primer.
■ Improper primer design, and the target sequence has homology with the non-target sequence. ——Re-design primers.
Phenomena: The PCR amplification bands are inconsistent with the expected size, either large or small, or sometimes both specific amplification bands and non-specific amplification bands occur.
A-1 Primer
■ Poor primer specificity
——Re-design primer.
■ The primer concentration is too high ——Properly increase the denaturation temperature and prolong denaturation time.
A-2 Mg2+ concentration
■ The Mg2+ concentration is too high ——Properly reduce the Mg2+ concentration: Optimize the Mg2+ concentration by a series of reactions from 1 mM to 3 mM with an interval of 0.5 mM to determine the optimal Mg2+ concentration for each template and primer.
A-3 Thermostable polymerase
■ Excessive enzyme amount ——Reduce enzyme amount appropriately at intervals of 0.5 U.
A-4 Annealing temperature
■ The annealing temperature is too low ——Appropriately increase the annealing temperature or adopt the two-stage annealing method
A-5 PCR cycles
■ Too many PCR cycles ——Reduce the number of PCR cycles.
A-1 Primer ——Poor specificity ——Re-design the primer, change the position and length of the primer to enhance its specificity; or perform nested PCR.
A-2 Template DNA
——The template is not pure ——Purify the template or extract DNA with purification kits.
A-3 Mg2+ concentration
——Mg2+ concentration is too high ——Properly reduce Mg2+ concentration: Optimize the Mg2+ concentration by a series of reactions from 1 mM to 3 mM with an interval of 0.5 mM to determine the optimal Mg2+ concentration for each template and primer.
A-4 dNTP
——The concentration of dNTPs are too high ——Reduce the concentration of dNTP appropriately
A-5 Annealing temperature
——Too low annealing temperature ——Appropriately increase the annealing temperature
A-6 Cycles
——Too many cycles ——Optimize the cycle number

The first step is to choose the appropriate polymerase. Regular Taq polymerase cannot proofread due to lack of 3’-5’ exonuclease activity, and mismatch will greatly reduce the extension efficiency of fragments. Therefore, regular Taq polymerase cannot effectively amplify target fragments larger than 5 kb. Taq polymerase with special modification or other high fidelity polymerase should be selected to improve extension efficiency and meet the needs of long fragment amplification. In addition, the amplification of long fragments also requires corresponding adjustment of primer design, denaturation time, extension time, buffer pH, etc. Usually, primers with 18-24 bp can lead to better yield. In order to prevent template damage, the denaturation time at 94°C should be reduced to 30 sec or less per cycle, and the time to rise temperature to 94°C before amplification should be less than 1 min. Moreover, setting the extension temperature at about 68°C and designing the extension time according to the rate of 1 kb/min can ensure effective amplification of long fragments.
The error rate of PCR amplification can be reduced by using various DNA polymerases with high fidelity. Among all the Taq DNA polymerases found so far, Pfu enzyme has the lowest error rate and the highest fidelity (see attached table). In addition to enzyme selection, researchers can further reduce PCR mutation rate by optimizing reaction conditions, including optimizing buffer composition, concentration of thermostable polymerase and optimizing PCR cycle number.
Products categories
WHY CHOOSE US
Since its establishment, our factory has been developing first world class products with adhering the principle
of quality first. Our products have gained excellent reputation in the industry and valuabletrusty among new and old customers..








